Multi-omics single-cell data integration and regulatory inference with graph-linked embedding
2022-05-04
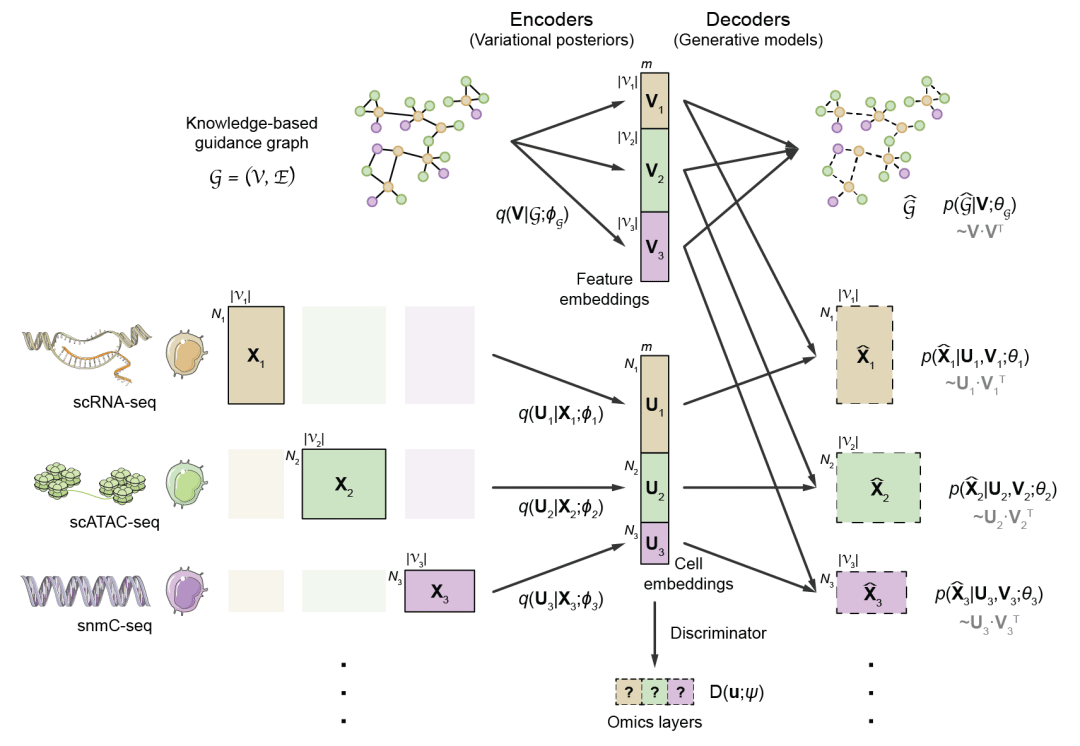
With rapid advances in recent years, single-cell omics technologies have become essential tools for studying cellular function and gene regulation. Up until now, many omics layers can be profiled at single-cell resolution, including transcriptome, chromatin accessibility, chromatin contact, DNA methylation, etc., each of which reflects one specific aspect of the cellular state. To achieve more holistic characterization of cellular states as well as the underlying regulatory circuits, multiple omics layers need to be analyzed in an integrative manner. While advanced experimental methods have been developed recently that can profile multiple omics layers within the same single cell, such methods are typically more complicated than their single-omics counterparts, and produce data of lower quality. More importantly, the combination of omics layers susceptible to simultaneous detection is limited, and it is unrealistic to profile all omics layers in the same cell.
Under this situation, computational integration of multiple unpaired single-omics datasets is of great significance, which can approximate multi-omics experiments to a certain extent without specialized experimental techniques, and potentially expand the boundaries of single-cell multi-omics analysis. The key challenge of computational multi-omics integration is the discrepancy in feature space among different omics layers. For example, the feature space of transcriptome consists of genes, while that of chromatin accessibility consists of open chromatin regions. Such discrepancy causes a lack of comparability between cells in different omics layers. Additionally, technological advances have dramatically increased the throughput of single-cell sequencing, now reaching up to millions of cells per dataset, posing serious computational challenge to large-scale data integration.
In order to address these challenges and achieve efficient and accurate single-cell multi-omics integration, on May 2, 2022, Dr. Ge Gao’s lab at Biomedical Pioneering Innovation Center (BIOPIC) at Peking University, Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI) of Peking University School of Life Sciences, and State Key Laboratory of Protein and Plant Gene Research, published a research article in Nature Biotechnology titled “Multi-omics single-cell data integration and regulatory inference with graph-linked embedding”, which officially releases the deep learning-based GLUE model for single-cell multi-omics data integration and regulatory inference.